- Programme Area 4
Immuno-Epigenetics

- Cell therapy
- Epigenetic imprinting
- Epigenetic Regulation
- T-Lymphocytes
Understanding the epigenome in T cells to develop new therapies
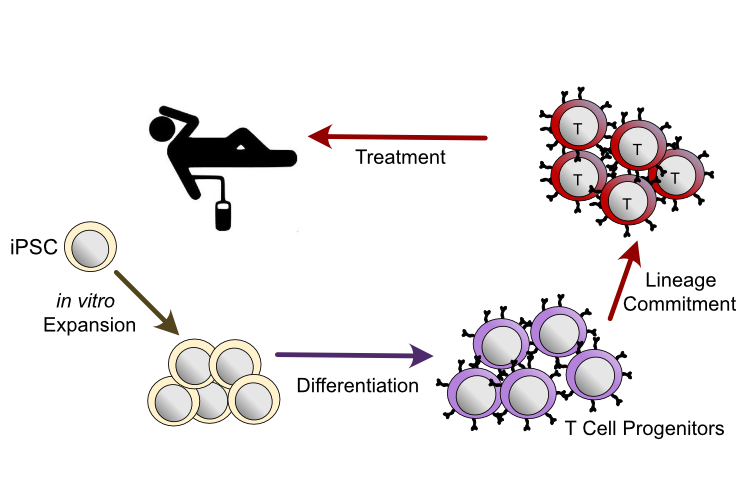
For many rheumatic diseases, curative therapies are still missing, leaving patients dependent on lifelong immunosuppressive treatment. Adoptive cell therapies offer a promising strategy for curative therapies with the goal of achieving full immunological homeostasis – either by permanently enhancing the immunosuppressive arm of the immune system (i.e. Treg therapy) or by eliminating the pathogenic autoreactive part (e.g. CAR T cell therapy).
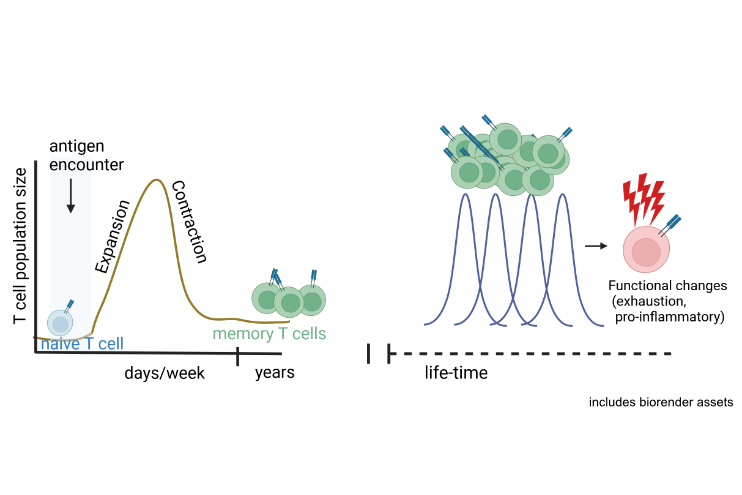
Our research group investigates the epigenetic mechanisms governing T cell differentiation, function, and senescence in both health and disease. From the knowledge gained, we aim at developing approaches for directed epigenetic interventions to generate new cell therapies for autoimmunity and chronic inflammatory diseases. With this, we make use of the regulatory potential, which is mediated by the 3-dimensional folding of the genome, to steer T cells towards optimal functional states for their use in cell therapy approaches.
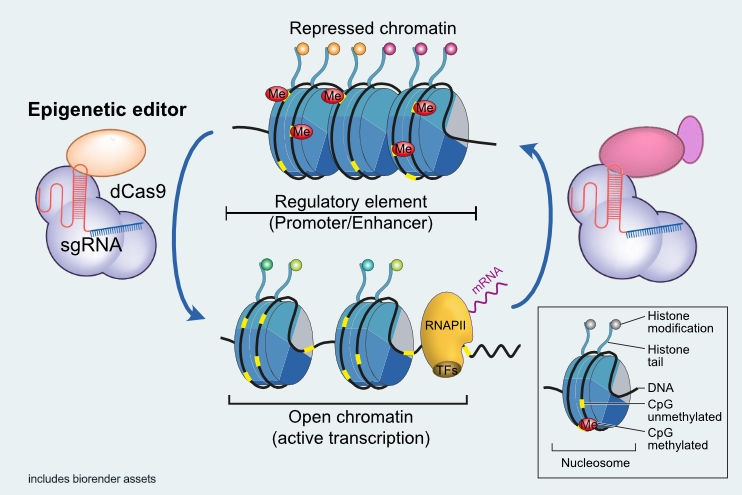
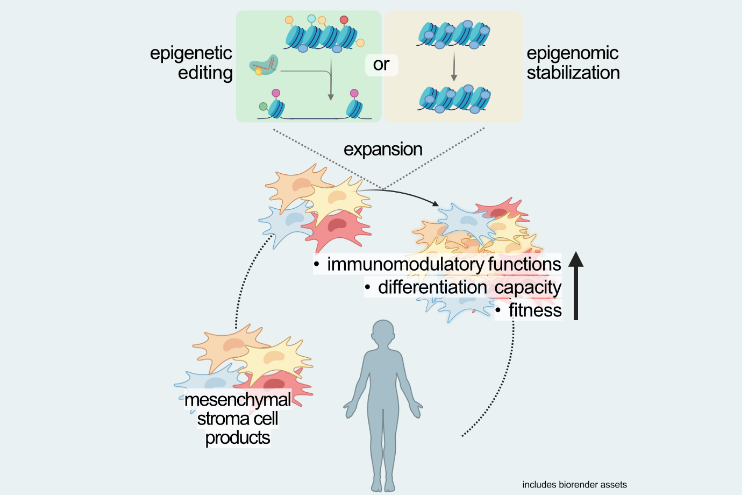
To this end, our group utilizes a broad portfolio of techniques for the epigenetic profiling of both, whole genomes and specific target genes, which are applicable to low input samples such as clinical specimen. In addition, our group develops and applies innovative approaches for the targeted modification of epigenomic structures, to be used to optimize therapeutic cell products.
By integrating fundamental epigenetic research with applied therapeutic strategies, our group seeks to advance the development of epigenetically engineered cell therapies for immune-mediated diseases.